Fish and microchips
Posted on 1 May 2019
Our researchers are gaining important new insights into the development of Parkinson’s by using the latest digital technology to study the way fish swim. It could take neurodegenerative research analysis to the next level.
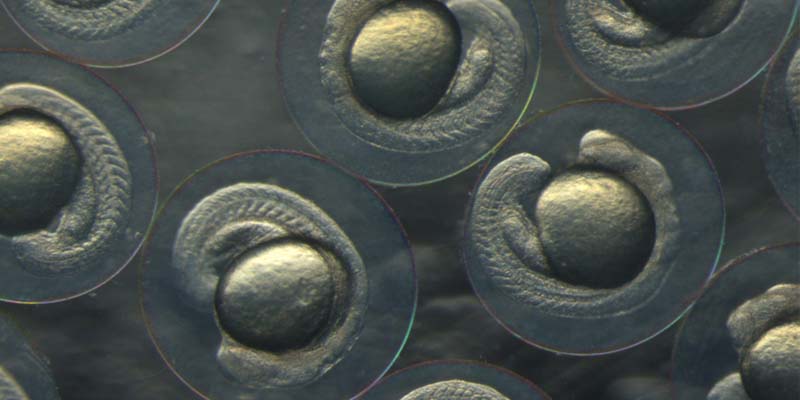
Zebrafish embryos.
We’re picking up differences in measurements which just couldn’t have been detected without this software. It’s early days and we need to be sure about our hypothesis on how tail movements relate to movements in humans, but we’re seeing some really exciting results.
To the uninitiated, humans may not appear to have much in common with the humble Zebrafish, but in many ways they’re remarkably similar, so when researching certain human diseases, they make an invaluable resource. Researchers at the University of York are currently using Zebrafish to model Parkinson’s and study the movement disorder.
Accurate
But when studying the movement of an animal, scientists want to be sure the data they’re extracting is accurate, and analysed objectively. Dr Betsy Pownall, of our Department of Biology, is leading a team which aims to bring digital accuracy and the objectivity of machine learning to its analysis.
She says: “Fish are vertebrates, like us, and they have neuroanatomy like ours, so when studying neurodegenerative conditions like Parkinson’s, we can see if the same degeneration occurs, so we can model the condition.”
Fish also have the advantage of laying eggs making their embryos easy to access in their early stages, and so manipulating genomes becomes easy - for a scientist at least.
The science behind Parkinson’s isn't straightforward, but Dr Pownall puts it simply: “In some cases Parkinson’s develops when certain genes become inactive - known as autosomal recessive Parkinson’s,” she says. “In other cases the condition might develop through overactive genes, but it was the recessive types I wanted to concentrate on for this project, because gene deletion methods work particularly well on them.”
Dr Pownall chatted with Professor Stephen Smith from our School of Physics, Engineering and Technology, whose own research into tracking the progression of Parkinson’s using digital sensors and computer algorithms, presented an intriguing possibility in her mind.
“We were talking about Stephen’s research in an airport bar, en route to a university event overseas,” she said. “I asked him if he thought his methods would work on fish.”
Professor Smith said he thought they probably would, and the idea to digitally monitor Zebrafish when researching Parkinson’s was born.
The first thing to be done was raise the fish and genetically alter them to develop the condition. EPSRC funded PhD student Gideon Hughes took the project on and identified a number of genes which, if lost, could cause the brain to develop Parkinson’s. He selected five such genes and set about raising genetically-altered, Parkinson’s-susceptible Zebrafish.
Capture
The next stage was to work out how to capture accurate information about the movements of the fish - and compare the movements with wild non genetically-altered fish. The answer lay in technology loved by outdoor enthusiasts and adventurers. “You can’t put a sensor on a fish, so I knew we would have to work with cameras,” said Gideon. “We wanted something waterproof that could record at a high frame rate, so we rigged up a 100 frame per second GoPro.”
Once the team had their film, they enlisted the help of Matt Bedder in the Department of Computer Science to create software which could track specific points along the spine of the fish, thus turning its movements into digital data. This data could then be fed into the evolutionary algorithms developed by Professor Smith to detect Parkinson’s in humans.
The beauty of this method says Professor Smith is that it removes any subjectivity, and the computer can discern even the slightest differences in movements which the human eye could never see. Also, the evolutionary nature of the algorithms means the software can learn which aspects of the fish’s movements are the most relevant in analysing the effects of Parkinson’s.
And although it’s early in the computational process, the team are excited about the results. Gideon is extracting measurements using the algorithms and comparing them against fish which don’t have the Parkinson’s-triggering genetic mutations.
Patterns
Gideon said: “Early findings are suggesting patterns in common with those we see in human patients. We’re picking up differences in measurements which just couldn’t have been detected without this software. It’s early days and we need to be sure about our hypothesis on how tail movements relate to movements in humans, but we’re seeing some really exciting results.”
And although the team know it’s a long way off, their hope is that the method could one day be used to find treatments for Parkinson’s disease.
Dr Pownall said: “In an ideal world we’d want to be able to treat these fish. So we’d compare a Parkinsonian fish to a normal fish using this method of digital analysis, then treat the Parkinsonian fish with a chemical; if the treated fish shows improvement in subsequent analysis then we know that chemical might be a candidate for Parkinson’s treatment in humans.”
- Read more about Professor Stephen Smith’s Parkinson’s monitors.
- Data analysis was carried out with the assistance of researchers at Heriot-Watt University.
The text of this article is licensed under a Creative Commons Licence. You're free to republish it, as long as you link back to this page and credit us.

Professor Stephen Smith
Research interests centred on the study of alternative representations for evolutionary algorithms and their application to problems in medicine and image processing.

Dr Betsy Pownall
Research Title: Reader, Department of Biology.
Research interests include the use of non-mammalian vertebrate embryos to study developmental mechanisms. The models used in Betsy's labs are Xenopus laevis and tropicalis and the Zebrafish, Danio rerio.
Explore more research
A research project needed to spot trees on historic ordnance survey maps, so colleagues in computer science found a solution.

We’re using gaming technology to ensure prospective teachers are fully prepared for their careers.

A low cost, high-accuracy device, could play a large part in the NHS's 'virtual wards'.
