Using yeast to dissect disease pathways
Lead researcher: Dr Chris MacDonald, Department of Biology
Cell surface membrane proteins play key roles in a broad range of physiological processes and form the largest class of approved drug targets. Despite this, fundamental questions regarding their regulation remain unanswered. Surface membrane proteins are regulated by the intracellular trafficking pathways that facilitate their transport to and from the plasma membrane, often traversing different organelles by vesicular transport.
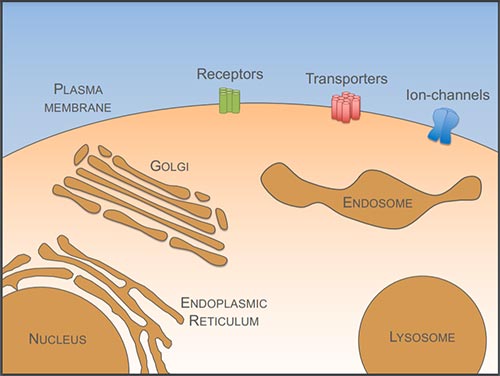
Figure 1: Cell surface membrane proteins perform a wide array of functions that are critical for cellular health and viability, such as receptors, nutrient transporters, and ion-channels. Surface proteins are synthesised at the endoplasmic reticulum and traffic to the surface via the Golgi apparatus. Surface proteins internalised to endosomes can recycle back to the plasma membrane or be sorted into the multivesicular body pathway for degradation in the lysosome.
Many features of these organelles and the pathways that connect them are evolutionarily conserved. We take advantage of this to understand mechanistic details of surface protein trafficking using the model organism, budding yeast. One advantage of yeast is powerful genetic tools, which we use to perform genome-wide screens to discover novel machinery related to surface protein trafficking.
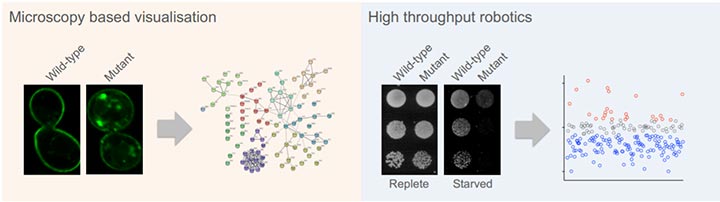 Figure 2: Using genome-wide collections we can systematically assess all yeast genes for specific roles in surface protein trafficking. By engineering a GFP-tagged reporter (left) we discovered many novel factors that regulate endosomal recycling of surface proteins [1]. By developing a growth-based assay that report on surface protein activity specifically in starvation conditions (right), we identified many factors that increase and decrease protein trafficking to the plasma membrane [2].
Figure 2: Using genome-wide collections we can systematically assess all yeast genes for specific roles in surface protein trafficking. By engineering a GFP-tagged reporter (left) we discovered many novel factors that regulate endosomal recycling of surface proteins [1]. By developing a growth-based assay that report on surface protein activity specifically in starvation conditions (right), we identified many factors that increase and decrease protein trafficking to the plasma membrane [2].
Following this discovery-based work, we have uncovered a series of mechanisms, involving highly conserved factors, that regulate trafficking of cell surface membrane proteins.
Understanding endosomal organisation in eukaryotes
Our work suggests that yeast surface proteins, much like mammalian counterparts, internalize to compartments that can be temporally classified as ‘early’ endosomes and functionally classified as ‘recycling’ endosomes [3]. Recycling from these compartments back to the plasma membrane is triggered by cargo deubiquitination [3,4,5] and appears to be distinct from recycling via the Golgi that relies on cargo ubiquitination [6,7].
Elucidating novel molecular mechanisms of endosomal recycling
We find that recycling of integral membrane proteins and lipids is regulated by a series of molecular mechanisms that control surface protein trafficking at endosomes.
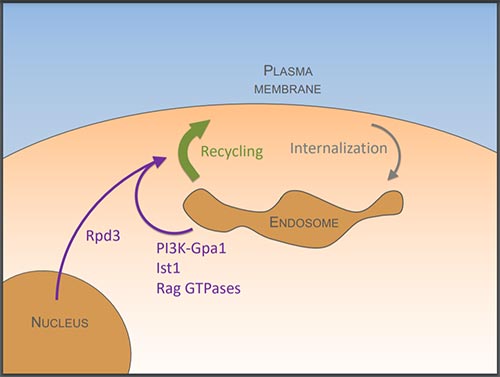
Figure 3: We show that endosomal lipids required for efficient cargo recycling are controlled by phosphatidylinositol 3-kinase (PI3K) and its downstream effector, the G⍺ subunit, Gpa1 [8]. This pathway also relies on the ESCRT associated factor Ist1, which we propose is involved in generating carriers for recycling [3]. Recycling is also regulated at the metabolic level via the Rag GTPases [1] and at the transcriptional level via the histone modifying Rpd3-complex [9].
Revealing how trafficking pathways are modulated during starvation
We are particularly interested in changes to the surface protein landscape in response to nutrient stress, which has large scale cellular effects, such as modulating molecular crowding [10,11]. As a cell survival mechanism, surface proteins are directed the lysosome for degradation in response to glucose starvation, and this trafficking is controlled by both an increase in traffic from the surface and a decrease in endosomal recycling back to the surface.
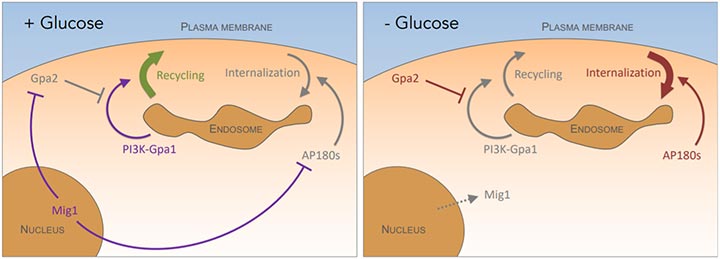 Figure 4: In glucose replete conditions (left), the Mig1 transcriptional repressor attenuates clathrin mediated endocytosis by repressing the AP180 proteins [12] and reduces recycling by repressing the Gpa2 inhibitor of PI3K-Gpa1 activity [8]. During glucose starvation conditions (right), the Mig1 repressor translocates from the nucleus resulting in increased AP180-mediated internalisation and inhibition of PI3K-Gpa1 recycling via Gpa2.
Figure 4: In glucose replete conditions (left), the Mig1 transcriptional repressor attenuates clathrin mediated endocytosis by repressing the AP180 proteins [12] and reduces recycling by repressing the Gpa2 inhibitor of PI3K-Gpa1 activity [8]. During glucose starvation conditions (right), the Mig1 repressor translocates from the nucleus resulting in increased AP180-mediated internalisation and inhibition of PI3K-Gpa1 recycling via Gpa2.
Nutrient transporters are regulated in subdomains of the plasma membrane
In addition to the membrane trafficking pathways that control protein movement to and from the surface described above, nutrient reporters are also regulated by localisation within subdomains of the plasma membrane itself. In response to nutrient starvation, a small reserve pool of nutrient transporters is sequestered in subdomains termed eisosomes, and this provides a physiological benefit to cells, promoting more efficient recovery from starvation [12].
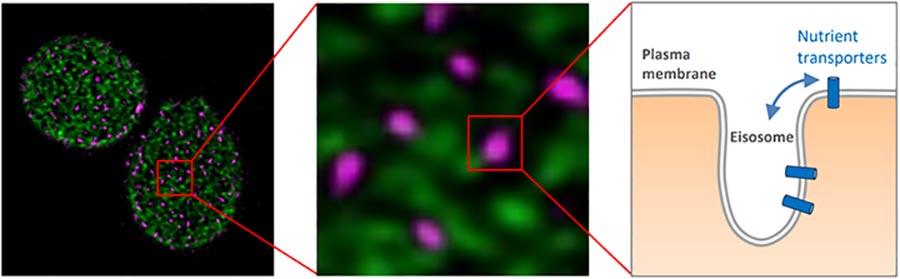
Figure 5: Eisosome compartments (pink) can be spatially distinguished from proteins is distinct subdomains (green). Nutrient transporters diffuse in and out of eisosomes (right).
References
- MacDonald C & Piper RC (2017). Journal of Cell Biology
- Paine KM, Ecclestone G, MacDonald C (2021). Traffic
- Laidlaw KM, Calder G, MacDonald C (2021). BioRxiv
- MacDonald C, et al. (2012). EMBO Reports
- MacDonald C, et al. (2015). Developmental Cell
- MacDonald C, et al. (2017). Traffic
- Xu P, et al. (2017). eLife
- Laidlaw KM*, Paine KM*, et al. (2022). Molecular Biology of the Cell
- Amoiradaki K*, Bunting KR*, et al. (2021). Int. Journal of Mol. Science
- Shepherd J*, Lecinski S*, et al. (2020). Methods
- Lecinski S*, Shepherd J*, et al. (2021). Current Topics in Membranes
- Laidlaw KM, et al. (2021). Journal of Cell Science
