Bacteria use selfish strategy to get ahead in the human gut
Posted on 21 October 2019
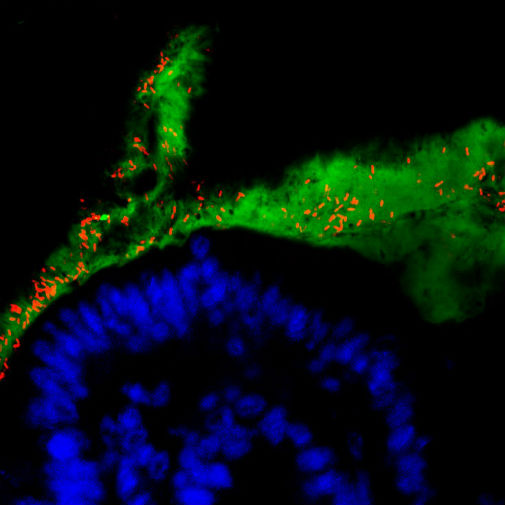
Bacteria (red) colonising the colon mucus layer (green). Image by Laura Vaux, the Quadram Institute
The findings could eventually lead to better diagnosis and treatment of conditions linked to imbalances in gut bacteria, the authors of the study say.
Ruminococcus gnavus (R. gnavus) is one of the trillions of microbes that call our digestive tract home. Known collectively as the ‘microbiota’ these microbes play a vital role in maintaining good health.
R. gnavus are one of several gut bacteria species that forage and feed on acidic sugars (called sialic acid) found in the protective mucus that lines the gut.
Source of nutrition
Led by researchers at the Quadram Institute in Norwich, and including scientists from the University of York, the research team found that while other mucus-foraging bacteria release a type of food called sialic acid for other members of the microbiota to metabolise, R. gnavus chemically modifies it so that only it can benefit.
The researchers discovered that R. gnavus has a protein that specifically transports the modified sialic acid into its cells. Once inside the cell, the bacteria can remove the modification, allowing them to metabolise the sugary acids as a source of nutrition.
The study revealed that the cluster of genes for this exclusive metabolic pathway are found in very few other bacteria. When the researchers knocked these genes out, it severely impaired the ability of the bacterium to colonise the mucus layer in the gut, indicating the importance of this process in allowing the bacteria to thrive.
Clever strategies
Professor Gavin Thomas, from the Department of Biology at the University of York, said: “Andrew Bell, from the Quadram Institute, visited our lab because of our leading expertise in how bacteria transport sialic acids.
“The findings of this study provide another great example of how bacteria competing with each other for limited food have evolved clever strategies to ensure they are not going to starve. It further demonstrates the complexity of the mixture of bacteria that lives in our gut and may also provide new targets for biomarkers or therapies for conditions linked to imbalances in the microbiota.”
R. gnavus is found in around 90% of humans and is considered to be a prevalent member of a ‘normal’ gut microbiota. However, it is also overrepresented in the microbiota of people suffering from a number of conditions including inflammatory bowel disease (IBD).
Next generation treatments
The new knowledge of how the bacteria gains a competitive advantage in the gut could aid the diagnosis of IBD and other conditions. It could also lead to next-generation prebiotic-based treatments targeting R. gnavus strains.
Lead author of the study, Professor Nathalie Juge from the Quadram Institute, said: “We think that by modifying the sialic acid, R. gnavus makes sure it always keeps a part of the cake for itself. This strategy contributes to its success in colonising the mucus layer and is likely to be behind its prevalence in the human gut microbiota.”
Explore more news
.jpg)
University of York awarded £1.1 million to lead the first stage of UK’s first comprehensive study on devolution and social security
Thursday 25 April 2024

More hybrid and nature-based solutions are urgently need to protect coastal communities from climate change
Wednesday 24 April 2024

New butterfly species created 200,000 years ago by two species interbreeding
Thursday 18 April 2024

Children in the North at greater risk of entering care
Wednesday 17 April 2024

Boreal forest and tundra regions worst hit over next 500 years of climate change, study shows
Monday 8 April 2024
Media enquiries
About this research
Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut is published in Nature Microbiology. This research was a collaboration between the Quadram Institute; the Universities of York, East Anglia and California; the Lerner Research Institute and Diamond Light Source. It was funded by the Biotechnology and Biological Sciences Research Council (BBSRC).
Explore more of our research.
