2D-IR Spectroscopy and Machine Learning Reveal Dynamic Protein Structures
Posted on Friday 9 January 2026
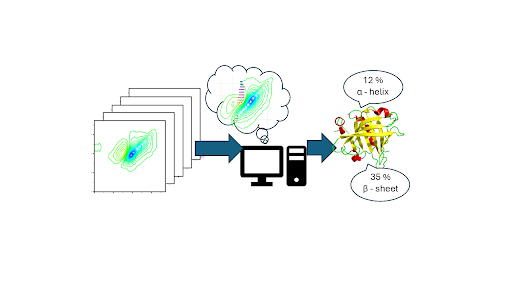
The dynamic three-dimensional structures of proteins define their biological function, but measuring these structures under physiological conditions is extremely difficult.
Ultrafast 2D-IR spectroscopy measures a unique protein structure fingerprint within minutes, using microlitres of label-free samples, in aqueous (H₂O) solution and with picosecond time resolution.
In a new paper published in Chemical Science, Professor Neil Hunt and members of his research team demonstrate that Machine Learning (ML) can convert these complex fingerprints into detailed structural information for the first time.
Using a library of 6732 2D-IR spectra of 35 proteins in H2O, the ML models were able to classify unknown protein samples according to structural content and measured quantities of α-helix and β-sheet accurately to within +/-7 %. The number and length of helices in a protein, and the presence of parallel and antiparallel β-sheets could also be predicted from the 2D-IR fingerprint.
These results come from the PhD project of Amy Farmer, which is jointly funded by EPSRC and The STFC Central Laser Facility. Amy said: “this work represents a key step towards rapid, quantitative analysis of dynamic protein structures under physiologically relevant conditions.”
** 2D-IR is available to external users as part of the Chemistry Analytical Facility
Notes to editors:
This work is published in the journal Chemical Science.
