Drug resistant infections/antimicrobial resistance
The excessive, inappropriate and often unnecessary use of antibiotics has dramatically accelerated the development of bacterial infections that are resistant to treatment. It is thus critical for clinicians to rapidly know which drugs are able to kill bacteria so they can select the right antibiotics. Current methods are inherently slow (24-48 hours) as they test for bacterial resistance by examining how bacteria grow in the presence of antibiotics.
We have been developing a range of innovative technologies that are capable of accelerating these bacterial susceptibility tests. For example, the primary mechanism of resistance against β-lactam antibiotics (one of the most important classes of antibiotics) is through the production of enzymes that can break down the antibiotics, rendering them ineffective. We have developed a modified form of a β-lactam antibiotic that can be attached to a sensor surface allowing us to rapidly detect the presence of these resistance enzymes. We have also demonstrated a miniature fluidic chip that allows us to trap hundreds of bacteria at once and monitor their response upon exposure to antibiotics. Critically, this microfluidic chip allows us to study antibiotic resistance of each individual bacteria. This not only improves the accuracy and speed of the test but also allows us to study the heterogeneity of resistance in bacterial communities.
This highly interdisciplinary research area involves a wide range of collaborators from across YBRI (including from the Departments of Physics (Krauss), Electronic Engineering (Johnson), Biology (Thomas) and Chemistry (Duhme-Klair) and with colleagues from the York Teaching Hospital NHS Foundation Trust.
L. M. Miller, C. D. Silver, R. Herman, A-K. Duhme-Klair, G. H. Thomas, T. F. Krauss, S. D. Johnson, Surface-Bound Antibiotic for the Detection of β-Lactamases, ACS Appl. Mater. Interfaces, 11, 32599 (2019)
G. Pitruzzello, S. Thorpe, S. Johnson, S., A. Evans, H. Gadêlha, T. F. Krauss, Multiparameter antibiotic resistance detection based on hydrodynamic trapping of individual E. coli , Lab on a chip, 19, 1417 (2019)
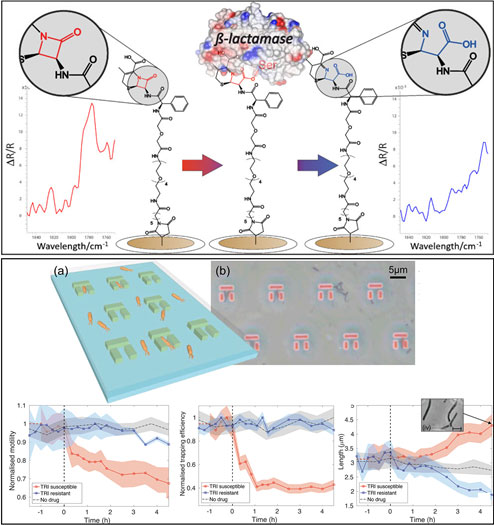
We have developed a surface-bound antibiotic (here cephalexin) which is compatible with the wide range of emerging surface-sensitive biosensing technologies and able to respond to the presence of β-lactamases,. Hydrolysis of the antibiotic by the presence of bacterial β-lactamases can be detected to indicate resistance to the drug. By hydrodynamically capturing hundreds of bacteria in microfabricated traps (a & b), we can simultaneously monitor the evolution of motility and morphology of individual bacteria upon drug administration to rapidly profile resistance.
