Molecular dynamics simulations: using physics to understand how biomolecules work
- By constructing appropriate force fields we can predict the motions of molecules at small, incremental times and describe their dynamics at atomic level
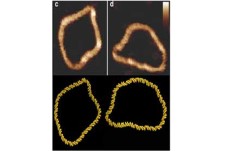
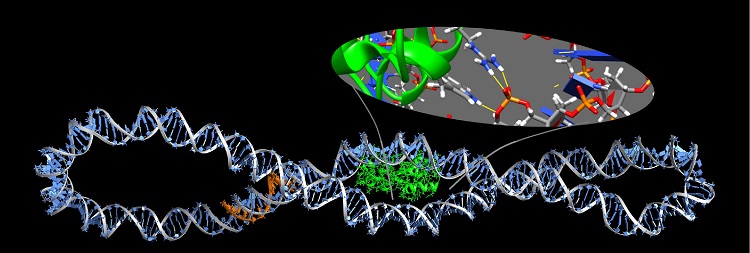
- These simulation tools give us huge insight into the shape of biomolecules and how these are influenced by solvent and other key molecules which bind to them
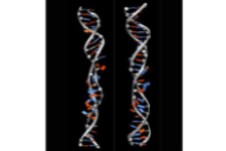
- The physical description of molecules like DNA, proteins or membranes, which are the building blocks of cells, is essential to understand the functioning of life
The Molecular Dynamics Simulation team is headed by Dr Agnes Noy, EPSRC Research Fellow and Proleptic Lecturer in Biophysics.

Dr. Noy is a computational biophysicist interested in the physico-chemical implications for the biological functionality of DNA and proteins. She obtained her degree in the University of Barcelona, and subsequently her PhD in theoretical and computational chemistry under the supervision of Prof. Modesto Orozco (IRB, Barcelona). In the next stage of her career, she moved to the UK with Prof. Ramin Golestanian at the University of Oxford by securing a “Beatriu de Pinos” and a EMBO Long-term postdoctoral fellowships. Her second postdoc was at the University of Leeds in collaboration with Dr. Sarah Harris and Prof. Tony Maxwell. She has gained research independence as an EPSRC Early-career Fellow and a Proleptic Lecturer in Biophysics at the University of York since 2016, leading the research theme of molecular dynamics simulations. Her research is centred in (i) the study of complex DNA topologies occurred by mechanical and torsional stress, (ii) the recognition of DNA by proteins and other ligands and (iii) the development of new computational tools for measuring how global flexibility emerge from little atomic fluctuations.
You can find more information on her Twitter @ANoyLab and on her webpage.