DNA elasticity
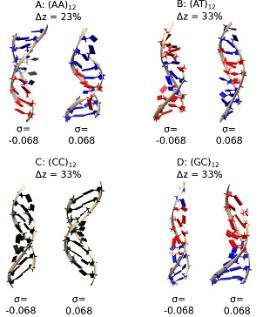
The double-helical structure of DNA results from canonical base-pairing and stacking interactions. However, variations from steady-state conformations result from mechanical perturbations in cells. These different structures have physiological relevance but their dependence on sequence remains unclear. Here, we use molecular dynamics simulations to show that sequence differences result in markedly different structural motifs upon physiological twisting and stretching [1]
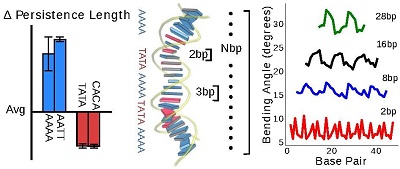
The resistance of DNA to stretch, twist and bend is broadly well estimated by experiments and is important for gene regulation and chromosome packing. However, their sequence-dependence and how bulk elastic constants emerge from local fluctuations is less understood. We have designed SerraNA [2], which is an open software that calculates elastic parameters of double-stranded nucleic acids from dinucleotide length up to the whole molecule using ensembles from numerical simulations. The program reveals that global bendability emerge from local periodic bending angles in phase with the DNA helicoidal shape. We apply SerraNA to the whole set of 136 tetra-bp combinations and we observe a high degree of sequence-dependence with differences over 200% for all elastic parameters. AT-rich motifs can generate extreme mechanical properties, which are critical for creating strong global bends when phased properly. Our results also indicate base mismatches would make DNA more flexible, while protein binding would make it more rigid. This software is based on the Length-Dependent Elastic Model (LDEM) previously described [3] and, from this perspective, the model was applied to test the new amber bsc1 force-field Parmbsc1 [4]
Publications
- JW Shepherd, RJ Greenall, MIJ Probert, A Noy, MC Leake (2020). The emergence of sequence-dependent structural motifs in stretched, torsionally constrained DNA. Nucleic Acids Research, 48, 1748-1763. https://doi.org/10.1093/nar/gkz1227
- V Velasco-Berrelleza, M Burmann, JW Shepherd, MC Leake, R Golestanian, A Noy (2020). “SerraNA: a program to determine nucleic acids elasticity from simulation data” Phys Chem Chem Phys, 22, 19254-19266 https://doi.org/10.1039/D0CP02713H
- A Noy, R Golestanian (2012) Length dependence of DNA mechanical properties Phys Rev Lett, 109, 228101. DOI:10.1103/PhysRevLett.109.228101.
- II Ivan, PD Dans, A Noy, A Perez, I Faustino, A Hopsital, J Walther, P Andrio, R Goni, A Balaceanu, G Portella, F Battistini, JL Gelpi, I Gomez-Pinto, C Gonzalez, M Vendruscolo, CA Laughton, SA Harris, D Case and M Orozco (2016). Parmbsc1: a refined force-field for DNA simulations Nature Methods, 13, 55. DOI:10.1038/nmeth.3658
