Study uses computer modelling to identify ‘vulnerable sites’ on coronavirus protein
Posted on 6 January 2021
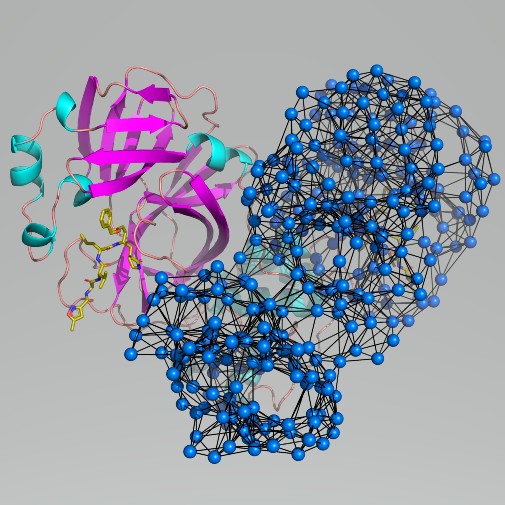 Scientists have used computer modelling to identify potential ‘vulnerable sites’ on a key protein found in coronavirus
Scientists have used computer modelling to identify potential ‘vulnerable sites’ on a key protein found in coronavirus
The coronavirus responsible for the Covid-19 epidemic deploys dozens of viral biomolecules when it invades host cells with the disease. One of these is a compact protein, the ’main protease’, whose function is critical to the virus.
Scientists at the University of York analysed the structure of the protease by computational modelling - simulating the protein’s motions.
New drugs
The model is very efficient because it works by developing a bird’s eye view of the protein structure. It suggests sites on the protein that may be accessible to new drugs.
The technique could be used for spotting ‘vulnerable’ sites for inhibition in other proteins, including other SARS-CoV-2 proteins.
The study, by Tom McLeish, Professor of Natural Philosophy in the Department of Physics, and Igors Dubanevics from the School of Natural Sciences, is published in the Journal of the Royal Society Interface.
Spike protein
Prof McLeish said the research was not related to the current vaccines, which are based on the ’Spike’ protein, but is a study of another key protein in the Covid process.
He added: “It is more relevant to potential future drugs than to future vaccines, as the motions of the protein that it uncovers point to new ’sites’ on the protein where binding small molecules might disrupt the protein function.
“The advantage of these sites, and our method in general, is that they are not the ‘obvious’ ones that compete with the normal binding of the protein, but other sites that can be accessed even when the usual binding sites are occupied.”
Simulations
Igors Dubanevics added: “We have identified promising druggable sites in the main protease via computer simulations and some of them have been supported by the newest studies by other groups.
“The next logical step would be to investigate the identified sites by conducting biological experiments in a lab.
Explore more news

Cells that communicate may help hinder bladder cancer from spreading
Thursday 19 February 2026

Police need better coordination on mental health emergencies, study shows
Thursday 19 February 2026

£2.5 million investment boosts pioneering cancer research
Wednesday 11 February 2026

UK growth ambitions at risk without more graduates to power priority industries
Sunday 8 February 2026

UK polling clerks struggle to spot fake IDs, study reveals
Wednesday 4 February 2026
Media enquiries
About this research
The new paper, which is freely available to download, is available here
Explore more of our research.
Our response to the coronavirus pandemic
We're working with partners in York and further afield as part of a global effort to fight the COVID-19 virus. From covid analysis in the labs to producing face shields for the frontline, we're using our knowledge and expertise to support the effort.
- See how we're helping the city and region
- Explore our COVID-19 research
