Dr Mahima Sharma
Research Fellow
Visit Dr Mahima Sharma's profile on the York Research Database to:
- See a full list of publications
- Browse activities and projects
- Explore connections, collaborators, related work and more
Profile
Biography
Mahima Sharma is a Research Fellow with expertise in structural enzymology and biocatalysis projects. She is currently working as a RCo-Investigator on BBSRC grant BB/W003805 on molecular dissection of sulfoglycolysis pathways with Professor Gideon Davies, FMedSci, FRS, at the York Structural Biology Laboratory.
Mahima is a 2021 RSC Horizon prize awardee for collaborative research with industrial partners, GSK, Prozomix, and Manchester Institute of Biotechnology and also won a recent 2024 RSC Horizon prize, for her work on breakdown pathways of ubiquitous plant sulfolipids, with researchers from University of Melbourne, Meiji University and Kyoto University.
Previously, she completed her DPhil in Chemical Biology from the University of Oxford in 2015 and worked on designing artificial metalloenzymes for C-C cross-coupling reactions under the supervision of Professor Benjamin G. Davis, FRS, and then began her postdoctoral work on structural and biochemical studies of enzymes enabling chiral amine synthesis (IREDs and reductive aminases) with Professor Gideon Grogan and Professor Nicholas J. Turner.
Research
Overview
Research interests
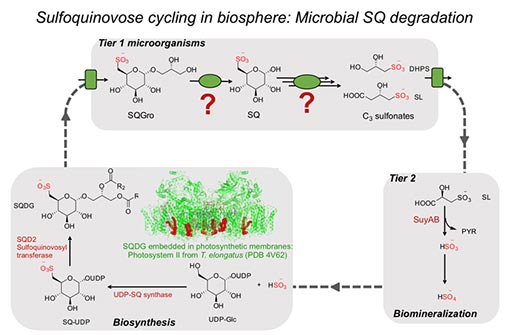
Her current project is focused upon investigation of degradation pathways of sulfoquinovose (SQ), a sulfur containing sugar liberated from sulfolipids found in thylakoid membrane of chloroplasts in plants. These SQ catabolic pathways prevalent in the environment will further our understanding of how sulfur is circulated from this major organosulfur reservoir.
Publications
Selected publications
- Detection of Sulfoquinovosidase Activity in Cell Lysates Using Activity-Based Probes
Z. Li†, I. Pickles†, M. Sharma† et al, Angew Chem Int Ed Engl, 2024, :e202401358. [†equal contribution] - Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases
M. Sharma et al, JBC, 2023, 299, 11, 13, 105338. - Widespread Family of NAD+-Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism
A. Kaur, I. B. Pickles, M. Sharma, N. M. Soler, N. E. Scott, S. J. Pidot, E. D. Goddard-Borger, G. J. Davies & S. J. Williams, JACS, 2023, 145, 51, 28216. - Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum
J. Li, M. Sharma et al, Chem Sci, 2023,14, 11429. - Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria
M. Sharma et al, PNAS, 2022, 119 (4) e2116022119. - Sulfoglycolysis: catabolic pathways for the breakdown of sulfoquinovose
A. J. D. Snow, L. Burchill, M. Sharma*, G. J. Davies*, S. J. Williams*, Chem Soc Rev, 2021, 50, 13628 [*corresponding author]. - The molecular basis of Sulfosugar selectivity in Sulfoglycoysis
M. Sharma et al, ACS Central Science, 2021, 7, 476. - Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
L. R. Rapp, S. M. Marques, E. Zukic, B. Rowlinson, M. Sharma, G. Grogan, J. Damborsky and B. Hauer, ACS Catal, 2021, 11 (5), 3182. - Dynamic Structural Changes Accompany the Production of 2-Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
M. Sharma et al, ACS Catal, 2020, 10 (4), 2826. - Asymmetric synthesis of primary amines catalyzed by fungal reductive aminases
J. Mangas-Sanchez & M. Sharma et al, Chem Sci, 2020, 11, 5052-5057. - Inverted binding of non-natural substrates in strictosidine synthase leads to a switch of stereochemical outcome in enzyme-catalyzed Pictet-Spengler reactions
E. Eger, A. Simon, M. Sharma, S. Yang, W. B. Breukelaar, G. Grogan, K. N. Houk, and Wolfgang Kroutil, JACS, 2020, 142, 792. - A Mechanism for Fungal Reductive Aminases (RedAms)
M. Sharma & J. Mangas-Sanchez et al, ACS Catal, 2018, 8, 12, 11534. - Biocatalytic Routes to Enantiomerically Enriched Dibenz[c,e]azepines
S. P. France, G. A. Aleku, M. Sharma, J. Mangas-Sanchez et al, Angew. Chem. Int. Ed., 2017, 56, 15589. - A Reductive Aminase Enzyme from Aspergillus oryzae
G. A. Aleku, S. P. France, H. Man, J. Mangas-Sanchez, S. L. Montgomery, M. Sharma, F. Leipold, S. Hussain, G. Grogan & N. J. Turner, Nature Chemistry, 2017, 9, 961. - NAD(P)H-Dependent Dehydrogenases for the Asymmetric Reductive Amination of Ketones: Structure, Mechanism, Evolution and Application
M. Sharma et al, Adv. Synth. Catal, 2017, 359, 2011.

Contact details
Find out more:

