Professor Gideon Davies, FMedSci, FRS
Royal Society Ken Murray Research Professor
Email: gideon.davies@york.ac.uk
Structural Enzymology and Chemical Biology in Glycoscience
Research Summary
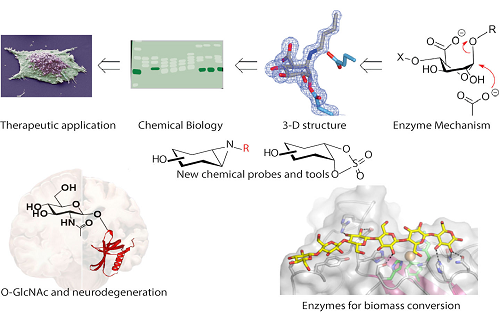
Our research spans the disciplines of Chemistry and Biology, with a focus on the structural enzymology and chemical biology of proteins involved in the synthesis and degradation of carbohydrates. Stimulated by the organic chemistry of enzymes, we develop and apply technologies to study the roles of glycans in living organisms with a keen eye to societal impact. Current projects include:
- Chemical enzymology of the sulfoquinovose sugar – a major source of biological sulfur
- Development and application of activity-based probes in the glycosciences
- Glycan biosynthesis and degradation in health and disease: viral invasion, cancer and imaging/treatment of lysosomal storage diseases
- Structure, function imaging and inhibition of heparanase; applications in cancer biology
- The O-GlcNAc modification and its role in neurodegeneration
- Dissecting the human intestinal microbiota and their role in glycan degradation
- Enzyme discovery, characterization and application in the field of plant polysaccharide degradation and of biomass conversion. The Cu chemistry and activity of fungal and bacterial lytic polysaccharide monooxygenases
Selected Recent Publications
- Modularly designed protein fragments combine into thousands of active and structurally diverse enzymes Lipsh-Sokolik et al., Science 2023, 379, 195-201.
- A multiplexing activity-based protein profiling platform for dissection of a native bacterial xyloglucan-degrading system McGregor et al., ACS Central Science. 2023 https://pubs.acs.org/doi/10.1021/acscentsci.3c00831
- A widespread family of oxidoreductive sulfoquinovosidases at the gateway to sulfoquinovose catabolism Kaur et al., J Am Chem Soc. 2023 in press
- Potent macrocyclic peptides that bind O-GlcNAc transferase inhibit glycosylation activity through an allosteric mechanism Alteen et al., Angew Chemie Int Ed 2023 e202215671
- Mechanism based heparanase inhibitors reduce cancer metastasis in vivo.
de Boer et al., Proc Natl Acad Sci USA 2022 119, e2203167119.- Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria Sharma et al., Proc Natl Acad Sci (USA) (2022) 119 e2116022119
- Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes.
Sabbadin et al., Science 2021 373, 774-779.- Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism Cuskin et al., Nature, 2015, 517, 165–169
- A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes
J Larsbrink et al., Nature, 2014, 506, 498-502
Biography
Gideon Davies received his PhD from the University of Bristol in 1990, and went on to postdoctoral research at EMBL Hamburg and CNRS Grenoble. In 1996 he received a Royal Society University Research Fellowship to work on Carbohydrate-Active Enzymes. He was made a Professor of the University of York in 2001. He has won many awards including the 2018 RSC Haworth Award, the 2016 iChemE Global Energy award, the 2015 Davy Medal of the Royal Society, the 2014 Khorana Prize of the Royal Society of Chemistry, and the 2010 Gabor Medal of the Royal Society. Gideon was elected a a Fellow of The Royal Society in 2010, and as a member of the European Molecular Biology Organization the same year. He was also elected a Fellow of the Academy of Medical Sciences in 2014. Gideon Davies was made a Royal Society Ken Murray Research Professor in 2016.

Find out more:
For up-to-date publication details please use the Google Scholar or "full publication list" links below.
Full publication list - Professor Gideon Davies (PDF
, 1,589kb)
Research funders include:
![]()

![]()
