Professor Neil C Bruce
Centre for Novel Agricultural Products (CNAP)
Research
Enzyme discovery for environmental and industrial biotechnology
Enzymes play a pivotal role in many biotechnological applications, including use as biocatalysts to make high value chemicals, in the saccharification of biomass for biofuel production and for environmental remediation of polluted sites. Recent developments in ‘omics technologies, bioinformatics and gene synthesis are providing powerful approaches to finding new and novel industrial enzymes from natural systems.
Research in Neil’s lab largely focuses on enzymes mediating plant and microbial metabolism of xenobiotic compounds. In particular, we have been studying the chemistry, biochemistry and molecular genetics of explosives metabolism. Explosive compounds 2,4,6-trinitrotoluene (TNT) and hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) used in munitions are highly toxic and the potential for progressive accumulation of such compounds in soil, plants and groundwater is a significant concern at military sites. We are characterising the enzymes involved in the detoxification and metabolism of explosives in plants and microbes and are using this knowledge to successfully engineer transgenic plants able to remediate toxic explosive pollutants. An innovative aspect of our work has been the use of synthetic biology to combine the biodegradative capabilities of explosives-degrading bacteria with the high biomass, stability and detoxification systems inherent in plants.

Transgenic switch grass remediating soil contaminated with the explosive RDX
Neil’s lab is also discovering new enzymes and associated proteins for biomass degradation. We have been using a multi ‘omics approach, combining the power of extracellular proteomics and transcriptomics, to identify proteins critical for lignocellulose deconstruction from microbial communities and animals obtained from marine and terrestrial environments. This approach is allowing the identification of new types of lignocellulose active proteins, both broadening our fundamental understanding of this process, as well as providing novel activities for research and industrial applications.
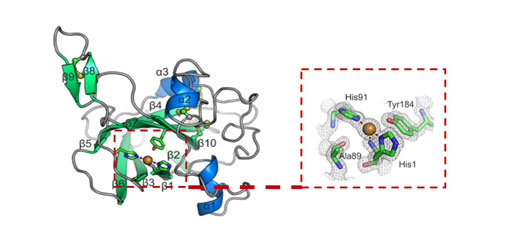
Structure of a lytic polysaccharide monoxygenase from Thermobia domestica
Teaching and scholarship
![]()
I particular enjoy teaching subjects related to biotechnology and my overarching goal is to demonstrate how the fundamental skills and knowledge students gain during their degree can be used to help solve real world problems. I believe critical thinking skills are important to success and encourage students to ask questions during lectures and query what is published in the literature. My experience has taught me that a well-formed question leads to a greater understanding of the subject. I draw on examples from my own research to highlight how fundamental knowledge in biology and biochemistry along with collaborations with scientists in other disciplines can help address environmental issues faced by industry and society.
![]()
My lectures generally focus on enzymes and their applications. Enzymes are remarkable biological catalysts that can often catalyse reactions that synthetic organic chemists only dream about. They can massively increase the rate of a reaction, they can display high specificity and can be regulated by other molecules. My lecture material covers fundamental enzymology, the role of cofactors in enzyme-catalysed reactions, their use as biocatalysts to make compounds of high value, such as pharmaceuticals, and as a means of removing some of the most recalcitrant pollutants in the environment.
![]()
My tutorials are loosely themed around environmental biotechnology, where students explore how biological processes can be applied to help alleviate some of the major global challenges we face. As part of these discussions, students get the opportunity to discuss topical issues, such as the use of genetically modified organisms in agriculture to generate biofuels and to clean-up the environment. Students give presentations on a chosen topic that forms the basis of group discussions, they learn to critically evaluate publications and get feedback on their written work.
![]()
Projects are carried out in the CNAP laboratories and are aligned with on-going research programmes being conducted by my lab group. Because of our broad research interests, the projects can involve plants, microbes, molecular biology, enzyme characterisation and many other techniques.


