Professor Fred Antson
01904 328255
Email: fred.antson@york.ac.uk
Protein-nucleic acid interactions
Research Summary
Our group's interests are in understanding the molecular events that control biological mechanisms and we are concentrating on protein - nucleic acid interactions because of their key role in many biological processes. The major aim is to study, by X-ray structural analysis combined with several biophysical/biochemical methods, the structure and function of protein-nucleic acid complexes present in molecular motors and in steady assemblies.
Mechanism of viral DNA translocation
The mechanism of DNA translocation by double-stranded DNA viruses, such as herpes viruses and tailed bacteriophages, is investigated using bacteriophage SPP1 as a model system. During viral particle assembly, the DNA is driven into a preformed procapsid through a portal protein (shown as a ribbon diagram above), a circular assembly which exists as a 13-subunit particle in its isolated forms or as a 12-subunit particle within the viral capsid. We aim to analyse the mechanism by which structural events associated with ATP hydrolysis by the viral ATPase lead to conformational changes within the portal protein; and the mechanism by which these conformational changes cause the DNA to translocate into the viral capsid. Our current model of structural changes in the portal protein during DNA translocation into a preformed viral capsid is shown in the movie. The portal protein‘s tunnel loops are shown as ribbons with one loop coloured in red; DNA is shown as a ball-and-stick model.
Protein-RNA interactions during transcription regulation
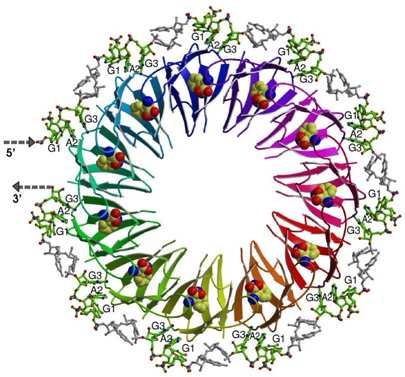
Here we use Bacillus subtilis system involving TRAP/anti-TRAP proteins and RNA as a model to study regulatory processes involving protein interactions with RNA sequences containing multiple repeated segments. The structure of TRAP/RNA complex (shown on Figure) explained the dependence of RNA binding on tryptophan. Recently, a protein of previously unknown function, now termed anti-TRAP, was shown to bind to TRAP and prevent it from interacting with RNA. We are now investigating the nature and antagonism of TRAP/anti-TRAP versus TRAP/RNA interactions.
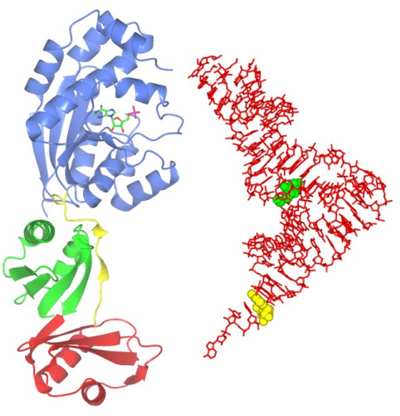
Structure and function of several tRNA modifying enzymes
Transfer RNA (tRNA) is a key element in the process of translation, whereby genetic messages are decoded in order to synthesise protein molecules. Cellular RNAs are frequently modified at the base or ribose moieties during maturation, and the greatest number and widest diversity of such modifications are found in tRNA. These modifications have various functions, including roles in the fidelity of translation and structural stability of tRNAs. As tRNA modifying enzymes are, in general, unrelated to each other, structural studies of members of each enzyme family are essential in understanding the molecular details of specificity and catalytic function. Using computational target selection strategies we have identified representatives of several such families and recently we determined the structure of Bacillus anthracis ThiI, a tRNA modifying enzyme performing a modification widespread amongst prokaryotic tRNAs. On the Figure, the ThiI protein complexed with AMP is shown to scale with the tRNA molecule (right). The target of modification by ThiI (uridine 8) is in green.
Selected publications
- Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids
Bayfield OW, Klimuk E, Winkler DC, Hesketh EL, Chechik M, Cheng N, Dykeman EC, Minakhin L, Ranson NA, Severinov K, Steven AC, Antson AA (2019) Proc Natl Acad Sci USA, 116, 3556-3561- Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Dehghani-Tafti S, Levdikov V, Antson AA, Bax B, Sanders CM (2019) Nucleic Acids Res. 47, 3208-3222.- Thermostable virus portal proteins as reprogrammable adapters for solid-state nanopore sensors.
Cressiot B, Greive SJ, Mojtabavi M, Antson AA, Wanunu M (2018) Nat Commun. 9, article 4652- S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Byrne RT, Whelan F, Aller P, Bird LE, Dowle A, Lobley CMC, Reddivari Y, Nettleship JE, Owens RJ, Antson AA and Waterman DG (2013) Acta Cryst D, 69, 1090-1098.- Structural basis for DNA recognition and loading into a viral packaging motor.
Büttner C, Chechik M, Ortiz-Lombardíaa M, Smits M, Ebong I-O, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson (2012) AA Proc. Natl. Acad. Sci. USA, 109, 811-816.- A flexible brace maintains the assembly of a hexameric replicative helicase during DNA unwinding.
Whelan F, Stead J, Shkumatov AV, Svergun DI, Sanders C, Antson AA (2012) Nucleic Acids Research, 40, 2271-2283.- Crystallographic snapshots of tyrosine phenol-lyase show that substrate strain plays a role in C-C bond cleavage .
Milic D, Demidkina TV, Faleev NG, Phillips RS, Matkovic-Calogovic D and Antson AA (2011) J. Am. Chem. Soc., 133, 16468-16476.- How to change the oligomeric state of a circular protein assembly: switch from 11-subunit to 12-subunit TRAP suggests a general mechanism.
Chen C-S, Smits C, Dodson GG, Shevtsov MB, Merlino N, Gollnick P and Antson AA (2011) PLoS One, 6, 1-11.- Structure and Activity of a Novel Archaeal beta-CASP Protein with N-Terminal KH Domains.
Silva APG, Chechik M, Byrne RT, Waterman DG, Ng CL, Dodson EJ, Koonin EV, Antson AA, Smits C (2011) Structure, 19, 622-632.- The crystal structure of unmodified tRNAphe from Escherichia coli.
Byrne RT, Konevega AL, Rodnina MV and Antson AA (2010), Nucleic Acids Research, 38, 4154-4162.
Career Summary
I graduated from the Moscow Institute of Physics and Technology (1986) and studied for PhD at the Institute of Crystallography (Moscow, USSR Academy of Sciences) in the laboratory of protein structure led by Boris Vainstein. During 1987-1989, as a member of a working group, I was involved in the USSR Academy of Sciences initiative to set up a synchrotron radiation station for protein crystallography at the Institute of Nuclear Physics (Novosibirsk, Russia). After a number of visits to European Molecular Biology Laboratory in Hamburg (1990-1992) where I worked with Keith Wilson, in 1992 I joined the laboratory of Guy Dodson at York. Since 1998 my research has been supported by the Wellcome Trust, this enabled me to set up a research group focused on studying structures and mechanisms of several protein-nucleic acid assemblies.
- 1998 - Wellcome Trust Research Career Development Fellowship
- 2002 – Wellcome Trust Senior Research Fellowship
- 2007 – Wellcome Trust Senior Research Fellowship, renewal for 5 years
- 2012 - Wellcome Trust Senior Research Fellowship, renewal for 5 years

