Dr Daphne Ezer
Lecturer in Computational Biology
Research
Computational questions
My research group primarily is interested in developing new computational methods for interpreting next generation sequencing data that comes from complex experimental designs, involving time series, combinatorial treatments, spatial analysis etc. We are interested in inferring the structure and interpreting gene networks, inferring gene-environment interactions, and studying the evolution of gene regulation in complex genomes. We are also interested in questions around 'heterogeneity' within populations of organisms or cells. We use statistics (modelling probability distributions, functional data analysis) and machine learning (supervised and unsupervised learning).
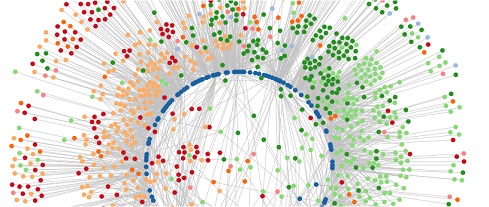
A snapshot of a gene network involving a large gene family of transcription factors.
Biological questions
Although the aim of the group is always to develop new methods that could be applied to many different kinds of research questions, the main biological focus of the group is around environmental signal integration in plants in the model organism Arabidopsis, but also sometimes extending to more tasty plants like Brassicas (a genus that includes cabbage, broccoli, turnips and Brussels sprouts) and Alliums (a genus that includes onions, leeks and garlic). For instance, some of our current projects address research questions such as: how do plants integrate light and temperature signals in the early morning to entrain the circadian clock? How has gene regulation evolved in complex (i.e. polyploid) Brassica genomes? How do spring onions respond to hourly temperature fluctuations?
Citizen science
In addition to classical approaches to research (laboratory experiments and lots of data analysis), we are also exploring alternative data collection strategies that involve engaging the public in the process of doing science. For instance, we have previously asked 50 primary schools to perform an experiment and collect data for one of our research projects. This allows data collections to be 'scaled up', while (i) minimising tedious measurement tasks for PhD students and postdocs and (ii) performing an engaging outreach activity.
Project supervision philosophy
I would like to provide supervision that is tailored for the needs of the student or postdoc. Every researcher would like independence and a sense of ownership in some aspects of the project, but might require lots of feedback and support in other phases of the project. I try to listen and figure out when to provide lots of one-on-one support and when to let the researcher fly free, exploring their creative ideas. I'm also a big fan of 'de-risking' a project, ie starting a supervisee on a clear sub-project that will definitely produce an outcome no matter what as long as effort is put in, but then having multiple opportunities for the student or researcher to extend the project in different directions based on their interests and skills.


