Dr Sangeeta Chawla
Senior Lecturer
Research
In mammalian neurons, synaptic activity-induced alterations in gene expression lead to enduring changes in synaptic transmission and cellular physiology that alter neuronal connectivity. These changes in neuronal connections are important for shaping neuronal circuits during development and for cognitive functions such as learning and memory. We study synaptic activity-dependent regulation of gene expression in cultured neurons from the rodent hippocampus, which is a region of the brain important for long-term memory formation.
Synaptic activity-dependent regulation of gene expression is triggered by the intracellular messengers Ca2+, cAMP and reactive oxygen species. We study the regulation of neuronal transcription factors by these messengers under physiological conditions, during normal brain ageing, and in neurodegenerative conditions.
|
In electrically active neurons HDAC4, a transcriptional corepressor protein, translocates to the cytoplasm. |
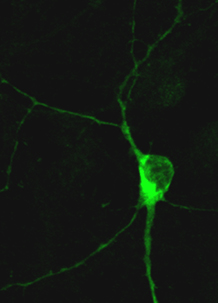 |
A major focus of the current work is to identify how synaptic-activity induced transcriptional changes in neurons influences their interaction with microglial cells and astrocytes. To do this, we are developing novel label-free imaging techniques that allow live imaging of neuronal-microglial interactions. View an example.
We are using Drosophila to investigate the role of conserved neuronal transcriptional regulators in learning and memory and in generating rhythmic behaviours such as circadian rhythms of locomotor activity. We have recently identified Class IIa HDACs as conserved regulators of circadian function in both fruit flies and mammalian cells. We are currently investigating the role of Class IIa HDACs in the altered sleep phenotype of flies modelling Parkinson’s.
|
The Class IIa HDAC, HDAC4 is required for robust circadian rhythms of locomotor activity in Drosophila. Double-plotted actograms of wild-type flies (control, left panel), which show robust patterns of daily activity, and HDAC4 hypomorphic flies (HDAC4KG09091, right panel), which are arrhythmic. |
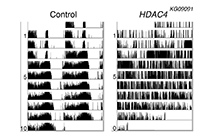 |
Teaching and Scholarship
![]()
I am a neuroscientist and teach primarily in this area. I take students through the scientific approaches and landmark studies that have helped unravel fundamental mechanisms and concepts with a view to helping students develop problem-solving skills applicable to any biological field.
![]()
I lecture in the Stage 2 Neuroscience module and run the Stage 3 Learning and Memory module. My lecture material aims to help students understand how synaptic transmission is modulated at the cellular level and how this underpins behaviours such as addiction, circadian rhythms and cognition. Evaluating the evidence from scientific studies for established knowledge and concepts is an important feature of my lectures.
![]()
My tutorial sessions for Stage 1 students aims to introduce them to ground-breaking work that has dramatically advanced biological understanding. This is done through critical discussions on Nobel prize-winning discoveries where students are free to choose a particular Nobel Prize or Nobel Laureate. For Stage 2 students I run tutorials on Cell Signalling where we consider how fundamental calcium signalling mechanisms are deployed in different cell types to mediate distinct physiological functions.
![]()
Undergraduate projects in the lab are aligned to our research interests in the timekeeping mechanisms of cellular circadian clocks. We investigate this in model organisms such as the fruit fly Drosophila melanogaster and the nematode Caenorhabditis elegans.


