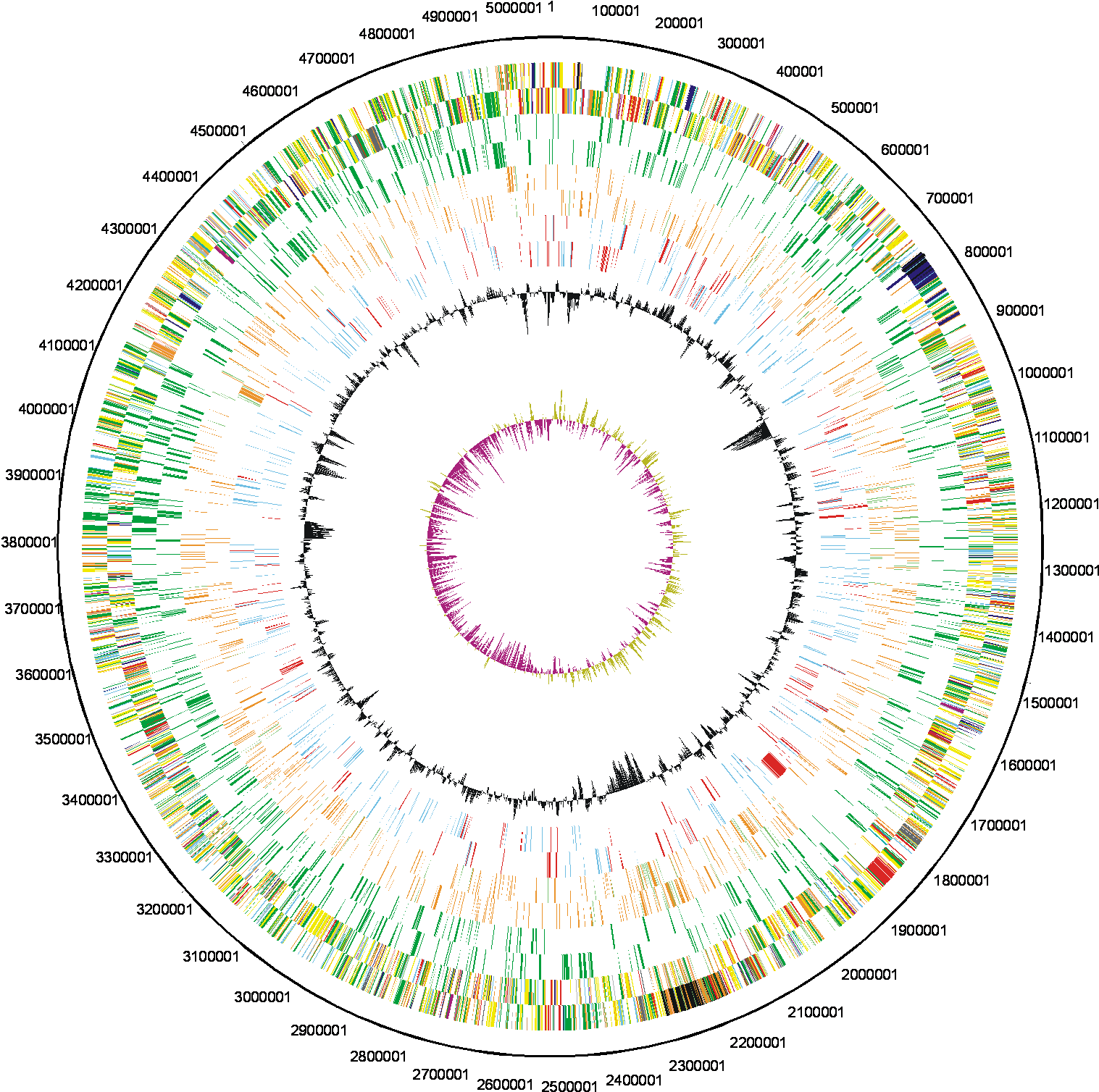
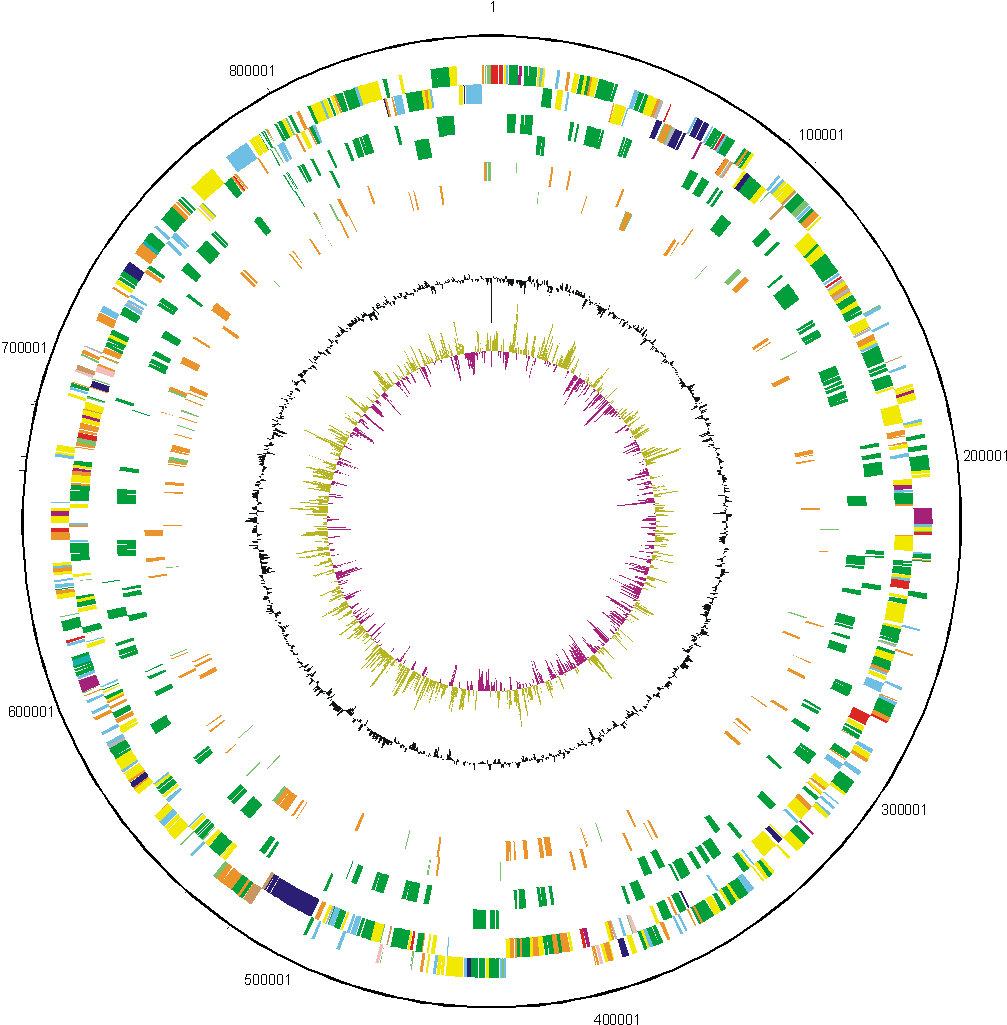
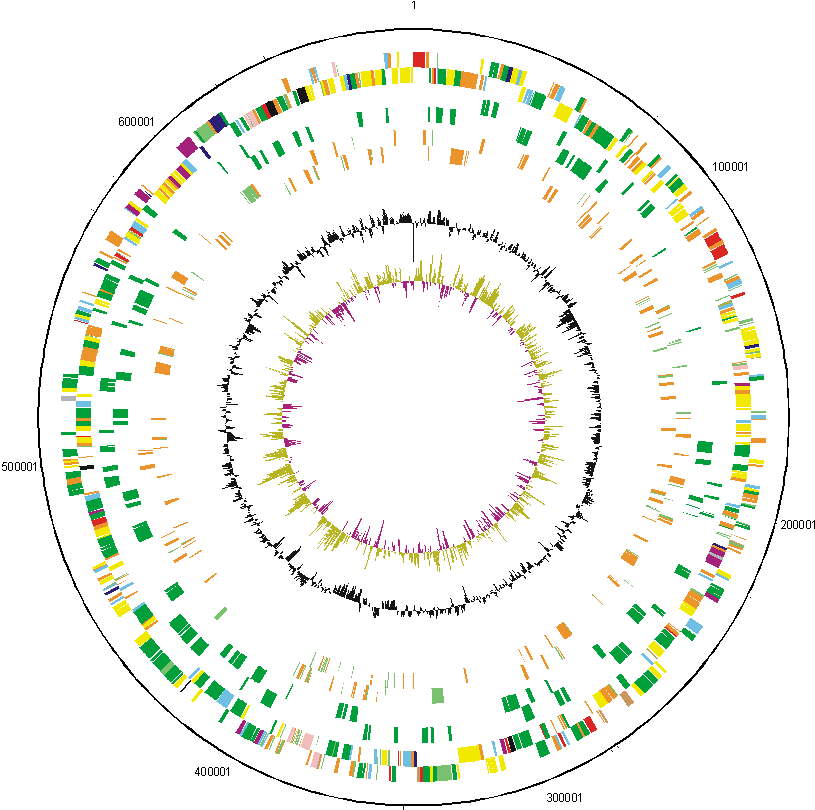
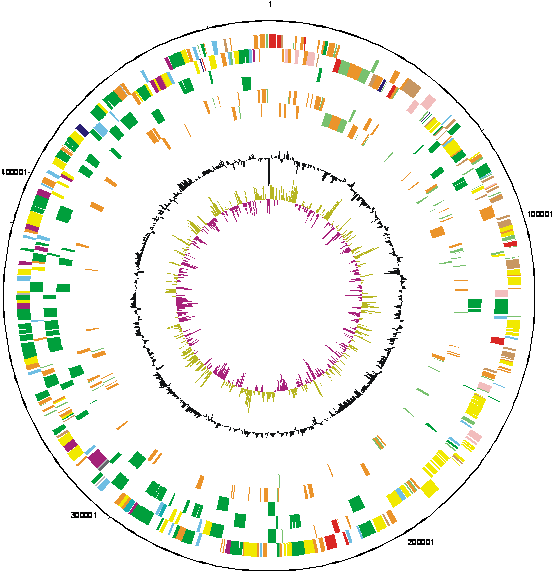
We have determined the complete genome sequence of Rhizobium leguminosarum biovar viciae strain 3841. The sequencing and annotation were carried out at the Sanger Institute, with funding provided by the Biotechnology and Biological Science Research Council (BBSRC). The award holders were:
Here are a few of the analyses extracted from that paper.
Genome
statistics
for Rhizobium leguminosarum biovar viciae
3841
|
Replicon
|
Base pairs
|
%G+C
|
Protein-coding
genes
|
Mean protein length
(a. a.)
|
rRNA operons
|
tRNA genes
|
|
Chromosome
|
5057142
|
61.1
|
4736
|
309
|
3
|
52
|
|
pRL12
|
870021
|
61.0
|
790
|
335
|
|
|
|
pRL11
|
684202
|
61.0
|
635
|
318
|
|
|
|
pRL10
|
488135
|
59.6
|
461
|
304
|
|
|
|
pRL9
|
352782
|
61.0
|
313
|
337
|
|
|
|
pRL8
|
147463
|
58.7
|
141
|
306
|
|
|
|
pRL7
|
151546
|
57.6
|
189
|
224
|
|
|
|
Overall
|
7751309
|
60.86
|
7265
|
309
|
3
|
52
|
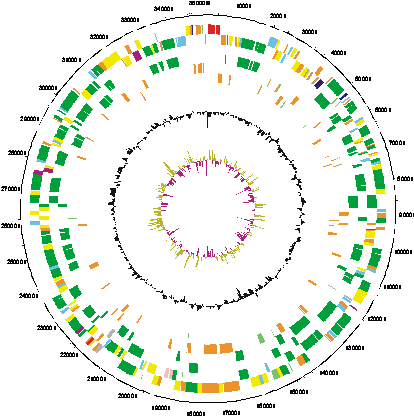
| Key to gene colours in the circular maps |
| 0 - white - Pathogenicity/Adaptation/Chaperones (shown here in dark blue) |
| 1 - dark grey - energy metabolism
(glycolysis, electron transport etc.) |
| 2 - red - Information transfer
(transcription/translation + DNA/RNA modification) |
| 3 - bright green - Surface
(IM, OM, secreted, surface structures[LPS etc]) |
| 4 - not used |
| 5 - turquoise - Degradation
of large molecules |
| 6 - pink/purple - Degradation
of small molecules |
| 7 - yellow - Central/intermediary/misc
metabolism |
| 8 - pale green - Unknown |
| 9 - pale blue - Regulators |
| 10 - orange/brown - Conserved
hypo |
| 11 - dark brown - Pseudogenes
and partial genes (remnants) |
| 12 - light pink - Phage/IS
elements |
| 13 - light grey - Some misc. infomation e.g. Prosite, but no function |
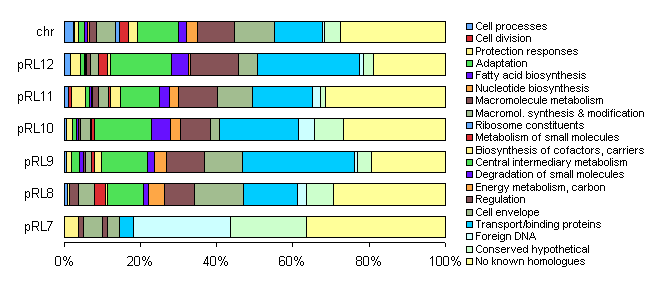 |
The distribution of functional classes of genes is fairly similar in the chromosome and larger plasmids, but pRL7 is very different. |
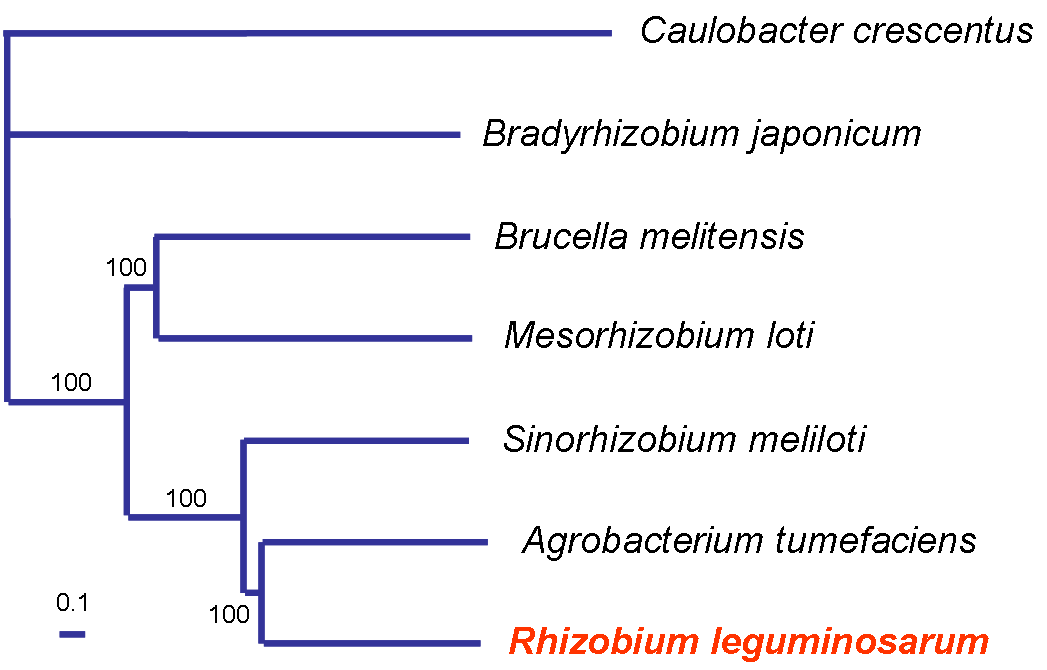 |
A phylogeny based on 648 orthologous protein sequences shared among seven alpha-proteobacterial genomes is similar to that based on small subunit ribosomal RNA |
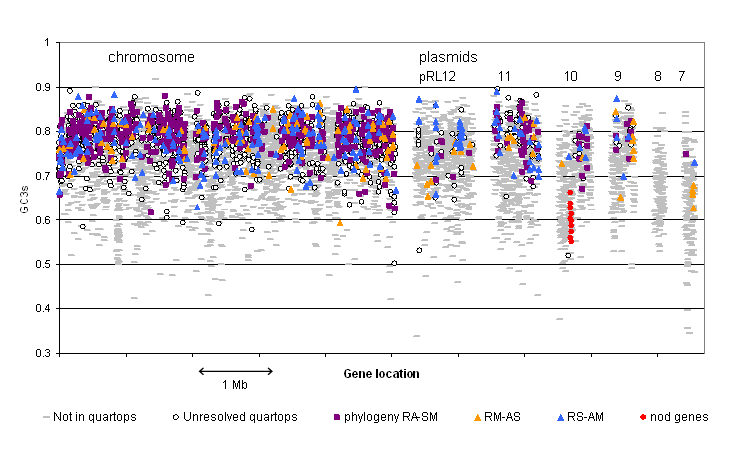 |
All protein-coding genes on
the chromosome and six
plasmids of R.
leguminosarum
3841, showing their nucleotide composition (GC3s: G+C content of silent
third positions of codons).Symbols
indicate whether
each gene encodes a protein with orthologs in A. tumefaciens,
S. meliloti
and M. loti, and, if so, which phylogenetic
topology it supports*.In
addition, the nodulation genes nodOTNMLEFDABCIJ are
identified on
pRL10.
|
| *RA-SM indicates a phylogeny that places Agrobacterium closest to Rhizobium, and so on |
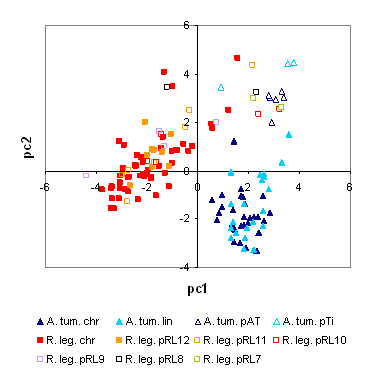 |
Dinucleotide compositional analysis of 100 kb regions of the genomes of R. leguminosarum 3841 and Agrobacterium tumefaciens C58, displaying the first two axes of a principal components analysis of the abundance of dinucleotides. Each chromosome (chr) and plasmid is identified by a distinct symbol. It is evident that the chromosome of each species has a different but consistent composition, while the smaller plasmids are more similar to each other. |
Almost all known rhizobia belong to the alpha-2 group of the Proteobacteria. Rhizobium leguminosarum is the type species of the type genus. It has three biovars - viciae, trifolii, phaseoli - that differ in their host specificity. Biovar viciae nodulates legumes in the Tribe Viciae - Vicia, Pisum, Lathyrus, Lens. The host-specificity genes are carried on large plasmids that also carry the genes for nitrogen fixation.
R. leguminosarum biovar viciae
strain 3841 has been widely
used in laboratory studies over the past 20 years. It is a spontaneous
streptomycin-resistant mutant of strain 300. It has a circular
chromosome
and 6 circular plasmids, ranging in size from approximately 147 to 870
kb. One of these plasmids (pRL10JI) carries the genes for nodulation
and
nitrogen fixation, but the function of the great majority of the
plasmid-borne
genes is unknown at present. All strains of R. leguminosarum
have
several large plasmids, but the number and sizes of plasmids varies
among
strains. This large number of plasmids distinguishes R.
leguminosarum
from the other rhizobia for which complete genome sequences have been
determined.
Mesorhizobium loti strain MAFF303099 has a large circular chromosome, into which the nodulation and nitrogen fixation genes are integrated as a "symbiosis island", and two circular plasmids. Its host plants are Lotus japonicus and related species. Note that a comparison of several basic genes suggests that this strain is more similar to the type strain of M. huakuii than to that of M. loti (TURNER, S L, ZHANG, X X, LI, F D, YOUNG J P W. 2002. What does a bacterial genome sequence represent? Mis-assignment of MAFF 303099 to the genospecies Mesorhizobium loti. Microbiology 148: 3330-3331.)
Sinorhizobium meliloti strain 1021 has a circular chromosome and two circular megaplasmids, one of which carries the nodulation and nitrogen fixation genes. Its host plants are alfalfa, Medicago sativa, and related species.
Agrobacterium
tumefaciens
strain C58 has a circular chromosome, a linear megaplasmid, and two
circular
plasmids. The smallest plasmid carries genes for tumour induction. This
species is not a rhizobium, in that it does not nodulate legumes or fix
nitrogen. Instead, it is a pathogen that causes stem galls on a wide
range
of plants. A. tumefaciens is, however, more
closely related to R.
leguminosarum than is either of the other two species, and
some taxonomists
favour its inclusion within the genus Rhizobium as R.
radiobacter.
Bradyrhizobium
japonicum strain 110 has a very large chromosome
(9.1 Mb) but no
plasmids. As in Mesorhizobium loti, the symbiosis
genes are in an
"island" of lower G+C content.
Brucella melitensis strain 16M and B. suis strain 1330 are animal pathogens, not rhizobia. They both have two small chromosomes (2.1 and 1.2 Mb). These strains are very similar in sequence and could be considered to belong to the same species. The larger chromosome shows extensive synteny with the chromosomes of Sinorhizobium meliloti and Agrobacterium tumefaciens.
Rhizobium etli is a close relative of R. leguminosarum, and the complete genome of strain CFN42 has been published.
A number of other genome projects for rhizobia have been completed or are under way, including the genomes of beta-proteobacteria that form root nodules on legumes, including Burkholderia phymatum STM815 and Cupriavidus taiwanensis LMG19424.
This page is maintained by Peter
Young at the University of York, and was updated on 8 April 2008.