Dr Gareth J O Evans
Senior Lecturer
Chair of the Board of Studies
Research
Protein Kinase Signalling in Neuronal Function
Protein phosphorylation is a rapid and reversible molecular switch used for intracellular signalling in diverse cell biological processes. The Evans lab studies the role of protein phosphorylation in a range of neuronal processes in the healthy and diseased brain, including development, learning, neurodegeneration and neuroblastoma.
The human genome encodes over 500 kinase enzymes, grouped in families that phosphorylate proteins within specific amino acid motifs. The effects of phosphorylation are diverse and include altering protein-protein interactions, enzyme activity or subcellular localisation. Identifying the specific substrates of each kinase is important because inherited and acquired mutations in kinases and their substrates are linked with a wide range of diseases, including cancer and neurological disorders.
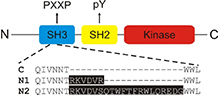
The Evans lab employs a variety of biochemical, proteomic, cell biological and fluorescence imaging approaches to study kinases and determine the function of their specific protein phosphorylation events. Our major goal is to determine the role of Src tyrosine kinase isoforms (C-Src, N1-Src and N2-Src) in the brain. Specific neuronal splicing events yield kinases that differ by just a few amino acids and yet have profoundly different functions. The N-Srcs have a role in neuronal development and their high expression is linked with a good prognosis in the childhood developmental cancer, neuroblastoma. We are investigating which N-Src substrates are linked with neuroblastoma prognosis and whether they provide a novel route to therapy.
|
Neuronal splicing of C-Src kinase yields N1- and N2-Src. Short inserts in the C-Src SH3 domain alter the kinase activity and substrate specificity of N1- and N2-Src. |
 |
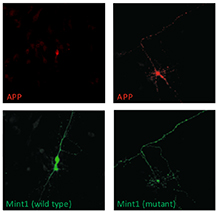
We also study the role of kinases in neurodegeneration. Aberrant trafficking of amyloid precursor protein (APP) is thought to contribute to the pathology of Alzheimer’s disease. We identified a single Src phosphorylation site in a trafficking protein, Mint1, which determines the destination of APP in neurons. We aim to establish if dysregulation of this switch-like mechanism is linked to the onset of Alzheimer’s disease.
|
A single phosphorylation site regulates APP trafficking. Wild type Mint1 confines APP to the cell soma (left panels) whilst mutation of its Src phosphorylation site permits APP to traffic throughout the neuron (right panels). |
 |
Teaching and scholarship
![]()
IThrough effective and entertaining teaching, I aim to enthuse students about the cellular and molecular mechanisms of life. I have extensive experience of programme and module design and the use of diverse methods of assessment. I am always keen to pilot new tools and techniques in the classroom. My teaching has been recognised by a Vice Chancellor's Teaching Award and two YUSU Excellence Awards for Innovative Use of Technology.
![]()
My broad background in biochemistry, neuroscience and intracellular signalling enables me to teach lectures, practicals and workshops on a variety of topics across all our degree programmes.
![]()
My tutorials often focus on neuroscience or signalling topics, but the main aim is to develop important transferable skills such as essay writing, understanding and criticising scientific papers, data interpretation and experimental design. Central to the success of my tutorials is providing timely and constructive feedback that I ensure students apply to their subsequent work.
![]()
Undergraduate and masters research projects offered in my lab use a combination of biochemical, cell biological and bioinformatic approaches to study neuronal signalling pathways relevant to brain development, dementia or neuroblastoma. Once familiar with conducting experiments and interpreting their data, students are encouraged to become more independent and take ownership of their research. Several of my project students have contributed data to Evans lab publications.



